Autism and autism spectrum disorders (ASDs) actually represent a rather large continuum of conditions that range from very severe neurodevelopmental delay and abnormalities to the relatively mild. In severe cases, the child is nonverbal and displays a fairly well-characterized set of behaviors, including repetitive behaviors such as “stimming” (for example, hand flapping, making sounds, head rolling, and body rocking.), restricted behavior and focus, ritualistic behavior, and compulsive behaviors. In more mild cases, less severe compulsion, restriction of behavior and focus, and ritualistic behaviors do not necessarily preclude functioning independently in society, but such children and adults may have significant difficulties with social interactions and communication. Because ASDs represent a wide spectrum of neurodevelopmental disorders whose symptoms typically first manifest themselves to parents between the ages of two and three, the idea that vaccines cause autism and ASDs has been startlingly difficult to dislodge and has fueled an anti-vaccine movement, both here in the U.S. and in other developed nations, particularly the U.K. and Australia. This movement has been stubbornly resistant to multiple scientific studies that have failed to find any link between vaccines in autism or the other favorite bogeyman of the anti-vaccine movement, the mercury-containing thimerosal preservative that used to be in many childhood vaccines in the U.S. until the end of 2001. Add to that the rising apparent prevalence of ASDs, and, confusing correlation with causation, the anti-vaccine movement concludes that vaccines must be the reason for the “autism epidemic.”
In reality, autism and ASDs appear to be increasing in prevalence due to diagnostic substition, better screening, and the broadening of the diagnostic criteria that occurred in 1994. Autism prevalence does not appear to be rising, at least not dramatically, at all, as the prevalence of ASDs, when assessed carefully, appears to be similar in adults as it is in children. If the true prevalence rate of autism and ASDs has increased, it has not increased by very much. In reality autism appears to have a major and probably predominant genetic component, and several scientific studies over the last few years have linked autism with various genetic abnormalities. Not surprisingly, given the varied presentation and severity of ASDs, these studies have not managed to identify single genes that produce autism or ASDs with a high degree of penetrance (probability of causing the phenotype if the gene is present). Indeed, one can argue that the state of current evidence is that ASDs are due to multiple genes, perhaps dozens or hundreds. Again, this is not surprising given the heterogeneity of ASD severity, presentation, and symptoms.
One of the more surprising studies supporting a genetic basis for autism appeared to much fanfare in Nature last week. The study by Pinto et al, looks at the functional impact of global rare copy number variation in autism spectrum disorders. Its results are rather surprising in that the large team of investigators (studies of this type take a lot of people to carry out) found that it may be relatively uncommon copy number variations in various genes that lead to the phenotype of autism or ASDs.
Before you can understand what this paper found, you need to know what copy number variation is. In basic genetics and biology classes, many of you probably learned, depending on how long ago you took the courses, that we have two copies of each gene, one on each chromosome, one inherited from the father and one inherited from the mother. The exception is that men have a Y-chromosome instead of a second X chromosome and therefore have only one copy of genes on the X and Y chromosome, the X-linked genes inherited from the mother, the Y-linked genes inherited from the father. Of course, nature is seldom quite so neat, and now that we have powerful tools for sequencing the entire genome and probing its entire DNA to seek out anomalies, we’re finding that things aren’t quite so simple. (Sometimes I marvel that we ever did think things to be that simple.) It turns out that there is considerable variation in the copy numbers of some genes in normal people. This is due to the rare duplication or deletion of a stretch of chromosome during replication. Such errors can leave a person with, for example, one copy on one chromosome and two on another. Although these events are rare for individual stretches of chromosomal DNA, over time and over many generations they can lead to some genes having several. Moreover, there appear to be “hot spots” in various chromosomes that are more prone to duplications or deletions than other regions of DNA.
Because I’m a cancer surgeon and biologist, I’m mostly familiar with variations in gene copy number associated with cancer, and there are many of these. Duplications and amplifications of stretches of DNA are practically the sine qua non of cancer. Often in cancer the amount of gene product in the form of protein that a given gene makes is proportional to the copy number. For instance, in breast cancer, there is a stretch of DNA known as the 8p11-12 amplicon. Basically, it’s a stretch of DNA on the p arm of chromosome 8 that is commonly amplified in breast cancer. This region of chromosome 8 is under active study, and there appear to be a number of candidate oncogenes there. One consequence of our learning about such amplified regions is that a formerly popular (about 30 years ago) and somewhat simplistic idea that single oncogenes would explain carcinogenesis, an idea that was later supplanted by a less simplistic but still too simple idea of multistage carcinogenesis in which a series of mutations in key oncogenes led to cancer. Now we understand that it is many genes that cause cancer. Whole networks of genes are usually perturbed, and, although there are commonalities in many of the networks purturbed among different cancers, the specifics can vary widely from cancer to cancer.
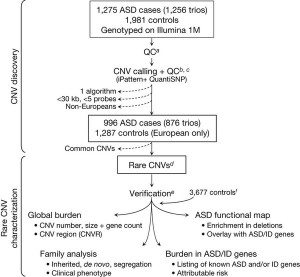
Cancer is one extreme, though, and obviously ASDs are not cancer (or, more appropriately, cancers, given that cancer is not just one disease). ASDs do, however, appear to require multiple genetic changes in order to manifest themselves. Although CNVs are associated with many diseases and conditions, the abnormalities in are not as dramatic or numerous as they are in cancer. In the study under discussion, Pinto et al undertook an a survey of as many CNVs as they could identify in autistic children. Basically, they genotyped 1,275 children with ASD and 1,981 neurotypical controls and compared the frequency of single nucleotide polymorphisms (SNPs) according to the following scheme:
It’s not necessary for you to understand in detail what SNPs are. Basically they are variations in single nucleotides within a gene that are useful because by examining them scientists can estimate CNVs and other genetic variants in various genes. This allows the performance of what is known as a genome wide association study (GWAS), which can look at genetic variation over the entire genome, which is what Pinto et al did using a technique that can look for SNPs in 1.2 million loci per sample. Genetic variations that are more common in people with a disease are said to be associated with that disease. GWAS can be very powerful, but, like much of modern genomic medicine, these studies produce incredible amounts of data, with all the attendant difficulties, both computational and scientific, in interpreting what all the associations that are found may or may not mean.
As the authors note in their introduction, previous attempts at GWAS for autism have identified candidate genetic loci at 5p14.1 and 5p15.2. However, variations in these loci can only account for a relatively small percentage of the heritibility of ASD, hence the desire to examine the entire genome in a large number of children with ASD.
What Pinto et al found was this:
The autism spectrum disorders (ASDs) are a group of conditions characterized by impairments in reciprocal social interaction and communication, and the presence of restricted and repetitive behaviours1. Individuals with an ASD vary greatly in cognitive development, which can range from above average to intellectual disability2. Although ASDs are known to be highly heritable (~90%)3, the underlying genetic determinants are still largely unknown. Here we analysed the genome-wide characteristics of rare (<1% frequency) copy number variation in ASD using dense genotyping arrays. When comparing 996 ASD individuals of European ancestry to 1,287 matched controls, cases were found to carry a higher global burden of rare, genic copy number variants (CNVs) (1.19 fold, P = 0.012), especially so for loci previously implicated in either ASD and/or intellectual disability (1.69 fold, P = 3.4 × 10-4). Among the CNVs there were numerous de novo and inherited events, sometimes in combination in a given family, implicating many novel ASD genes such as SHANK2, SYNGAP1, DLGAP2 and the X-linked DDX53-PTCHD1 locus. We also discovered an enrichment of CNVs disrupting functional gene sets involved in cellular proliferation, projection and motility, and GTPase/Ras signalling. Our results reveal many new genetic and functional targets in ASD that may lead to final connected pathways.
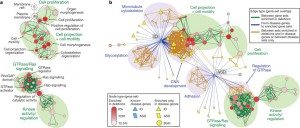
One interesting finding was that 5.7% of the CNVs discovered appeared not to be inherited; i.e., they were de novo, meaning, basically, new CNVs. More important, though, was the bioinformatic and systems biology analysis of the CNVs. If there’s one thing that the Human Genome Project has taught us it’s that single genes are rarely that important in disease. Usually, it’s genes that encode for various proteins in discrete functional classes and intracellular signaling pathways, something that can be analyzed by systems biology and network analysis, something I’m increasingly having to do for my cancer research. The result are “bubble diagrams,” which map out various networks and network “hubs” that are important in differences observed between normal and the condition being studied. To come up with such maps and pathway identification requires large numbers of specimens, terrabytes of computer storage space, and a lot of processing power, power that didn’t exist until this decade. In Pinto et al, such an analysis led to the identification of potential new pathways whose function in ASDs may be abnormal compared to the control group. These pathways are mapped out in the following illustration:
Candidate pathways for ASD identified by this method included genes involved with the cytoskeleton and microtubules, as well as genes involved in cell projection and motility, all of which are involved in cell migration. Other candidate pathways included genes involved in cell adhesion. It is not difficult to imagine how defective or altered migration and adhesion of neurons might result in the creation of abnormal neural pathways and thus result in the differences in cognition and behavior observed in ASDs. Of course, I’m putting things very simply. Even if this is true and these pathways are involved in ASDs, we know so little about how neural networks result in cognition and behavior that it will take a very long time and a lot of work to figure out exactly why and how abnormalities in these pathways result in ASDs. The same is true of other potential pathways implicated by these studies. These include GTPase and ras signaling pathways, as well as other kinase pathways. It’s not necessary for the lay reader to understand the full significance of these pathways. I don’t even know the full significance of these pathways in neurons, although I am familiar with them in cancer. Rather, it’s necessary only to know that they are involved in the transmission of signals from protein receptors on the cell surface into the cell, with the result being a number of processes, such as proliferation, migration, and the transmission of signals between neurons, among others.
Almost as important as the candidate pathways implicated by this study that clearly need further study to validate whether they are truly involved in the pathogenesis of ASDs or not are the pathways that were not implicated. One of the major claims of the “autism biomed” movement, the group of quacks who claim that they can treat autism with all manner of woo ranging from chelation therapy to various antioxidants and supplements, is that there are significant defects in pathways involved in countering the effects of oxidative stress, particularly pathways that result in glutathione production. (Glutathione is one of the major scavengers of reactive oxygen species–a.k.a. free radicals–in the cell.) Such claims were prominently featured by the lawyers for the complainants in the Autism Omnibus. Treatments allegedly targeting “detoxification” pathways involving “Glutathione, Cystathionine, Homocysteine, Methionine” figure prominently on the website of many a quack and are a favorite among the “vaccines cause autism” crowd. Don’t ask me how “vaccine injury” somehow causes oxidative stress sufficient to “cause autism.” Anti-vaccine “scientists” have long and convoluted pseudoscientific explanations that are implausible and unconvincing.
Guess what? Not a single one of the common “detoxification” or oxidative stress pathways implicated in ASDs by the “autism biomed” movement showed up in the analysis of the SNP data by Pinto et al. Big surprise, there. Well, not really. Of course, the fact that Pinto et al failed to find any of these pathways in ASDs in their analysis will no doubt be seized upon as “proof” that their analysis is hopelessly flawed. Just you wait. It’s coming. Because everything old is new again when it comes to the anti-vaccine movement I predict that Mark Blaxill will resurrect the same sorts of dubious criticisms of autism genetics studies that he made in this post from a year ago about an autism genetics study that predated Pinto et al:
But if the de novo CNV theory was plausible at one level, it was absurd at another. The genetic mutations the theory proposed (because this was the best the available evidence could support) were completely non-specific. The copy variants were spread widely (even randomly) over the genome, the theory went. No individual mutation was responsible for autism, just the unhappy presence of the wrong one. And these non-specific mutations were not only widely spread, they were virtually undetectable in the infant: no dysmorphic features; generally normal birth and (in many cases development); and the beautiful children we so often see affected by autism.
In other words these CNVs were a case of immaculate mutations.
And it was the perfect new project for the genetics research community. A wide open field of research opportunities. Lots of new money. And a chance to explain past failure away as part of the inexorable march towards genetic understanding.
Ah, yes. Likening science to religion. It’s a favorite canard of cranks of all stripes, be they anti-vaccinationists, creationists, alt-med promoters, 9/11 Truthers, and Holocaust deniers. I’ve heard anti-vaccine zealots refer to “Vaccinianity“; creationists refer to the “Church of Darwin“; and Holocaust deniers refer to “Holocaustianity” (warning: source is a French Holocaust denial website), among others. The intent is obvious: Try to paint the science detested as being faith-based rather than science-based.
The other favored attack, as those of you who read my post earlier today know, is to claim “conflicts of interest,” even when there aren’t any by any reasonable definition of the term. Although Blaxill hadn’t tried his hand at a pseudoscientific deconstruction of this study as of Sunday afternoon, John Stone over at the anti-vaccine propaganda blog Age of Autism has already pulled this gambit. In a post entitled Scherer of Nature Autism Gene Study Fails to Disclose Pharma Funding As Competing Interest, Stone opines:
Prof Stephen Scherer who is the senior author of the autism gene study launched in Nature last week holds the ‘GlaxoSmithKline-CIHR Pathfinder Chair in Genetics and Genomics at the Hospital for Sick Children and University of Toronto. The title used to be ‘GlaxoSmithKline-CIHR Endowed Chair”, GSK being one of the defendant companies in the UK MMR litigation
Mr. Stone is clearly ignorant of just what an endowed chair is. Basically, a company or wealthy donor gives a university a lot of money, and the university sets up an endowed chair using that money. The interest and dividends from the fund used to set up the chair are put at the disposal of the holder of the chair to do research and scholarship as he or she sees fit. The reason such chairs are desirable is because an endowed chair gives a researcher a reliable supply of funding without the need to write grant proposals or a department chair a source of funds for various projects that doesn’t have to come out of the departmental budget. It often allows a researcher to do more exploratory work or a department chair to engage in various research and educational activities by doling out funds from the chair to his faculty, for example, to support pilot projects by young faculty. Once an endowed chair is set up, the donor usually has no say over who gets the chair or how the money for the chair is spent. Claiming that Professor Scherer’s holding the GSK chair at his institution is an insurmountable COI that needed to be reported is, quite simply, ridiculous to anyone in academia who knows what an endowed chair is. Clearly, Mr. Stone does not although one commenter going by the ‘nym of Werdna does try to set Mr. Stone straight. It’s a rare thing indeed on AoA for a commenter to take a blogger to task like that, something that usually only happens when an AoA blogger makes a mistake so egregious even for some AoA readers.
Equally ridiculous is Mr. Stone’s lack of understanding of what a corresponding or first author is on a biomedical research paper:
While Prof Scherer’s departmental colleague Dalila Pinto is listed as lead author of the paper Scherer is listed as ‘correspondence author’ and he identifies himself as ‘senior author’ in Kevin Leitch’s LeftBrain/RightBrain blog.
Here’s a hint for Mr. Stone. The lead author of the paper is an honor usually reserved for the person who had the most to do with designing and doing the research. Most of the time the first author is the person who actually wrote the manuscript. On the other hand, the corresponding author of a scientific paper (at least a biomedical paper) is usually the author listed last or near last. More importantly for Mr. Stone’s criticism, the corresponding author and the senior author are nearly always one in the same, namely the author in whose laboratory and using whose funding the research described in the paper was performed. That’s it. That’s all those terms mean. There’s no inconsistency there. The terms are interchangeable. Mr. Stone would do well to learn a little bit about what he speaks before embarrassing himself so.
I’d be happy to educate him.
Mr. Stone’s follies aside, no doubt when Blaxill or whoever at AoA decides to attack Pinto et al shows up to do it, he’ll repeat the same breathtaking combination of ignorance and binary thinking that he did a year ago. As a pre-emptive rebuttal, I’ll cite popular science blogger P.Z. Myers, who put it well when he pointed out that these things are very complicated. So did Pinto et al:
Our findings provide strong support for the involvement of multiple rare genic CNVs, both genome-wide and at specific loci, in ASD. These findings, similar to those recently described in schizophrenia, suggest that at least some of these ASD CNVs (and the genes that they affect) are under purifying selection. Genes previously implicated in ASD by rare variant findings have pointed to functional themes in ASD pathophysiology. Molecules such as NRXN1, NLGN3/4X and SHANK3, localized presynaptically or at the post-synaptic density (PSD), highlight maturation and function of glutamatergic synapses. Our data reveal that SHANK2, SYNGAP1 and DLGAP2 are new ASD loci that also encode proteins in the PSD. We also found intellectual disability genes to be important in ASD. Furthermore, our functional enrichment map identifies new groups such as GTPase/Ras, effectively expanding both the number and connectivity of modules that may be involved in ASD. The next step will be to relate defects or patterns of alterations in these groups to ASD endophenotypes. The combined identification of higher-penetrance rare variants and new biological pathways, including those identified in this study, may broaden the targets amenable to genetic testing and therapeutic intervention.
The problem is that none of this sort of research is easy, and none of it is likely to result in effective treatments for ASDs very soon. Autism quacks and anti-vaccine zealots, however, can’t accept this. Again, like Blaxill, they demonstrate binary thinking. If a study doesn’t find a single, clear-cut gene or small set of genes causing autism or ASDs 100% of the time, then to them almost invariably the study is crap, autism is not genetic in origin, and vaccines (or other “environmental factors”–not vaccines, you know, nudge, nudge, wink, wink) cause autism. They also seem completely oblivious to developmental biology. It bothers them that a child with ASD appears normal at birth and then only manifests symptoms between the ages of 2 and 4. Anti-vaccine zealots who attack genetic linkage studies will frequently point this out as though this observation is a slam dunk argument against a genetic etiology for autism. Development proceeds, however, according to predictable, sequential steps that are under genetic control, and genetic variations and abnormalities can have a profound impact on development that may not manifest itself until previous parts of the developmental program are complete. For example, Tay-Sachs disease is one of thoes rare diseases for which a single gene is the known cause. Babies with Tay-Sachs usually appear normal at birth and develop normally for the first six months of life. Then the neurologic deterioration begins. There are also a number of inborn errors of metabolism that don’t manifest themselves at birth. In other words, it’s not unusual for purely genetic diseases to exhibit delayed manifestation of symptoms. Otherwise, why would we need to test for phenylketonuria (PKU) shortly after birth?
Antivaccine zealots also seem unable to understand that most chronic diseases and conditions with a strong genetic component are due to changes in activity of multiple genes, sometimes many genes. Due to functional redundancy in our cells, there are also often multiple abnormalities that can produce similar phenotypes; it is therefore not surprising that there might be many different gene abnormalities that contribute to ASDs. Systems biology gives us the tools to start to understand these exceedingly complex processes, as Pinto et al did in the study under discussion and as my collaborators try to do for cancer. The problem is that biology is really complex. Frustratingly to those waiting for treatments, it is far more complex than even scientists realized before the Human Genome Project and the dawn of genomic medicine.
While it is possible–even likely–that there is an environmental component to ASDs, the evidence to date has not convincingly implicated any, except for, ironically enough given how anti-vaccine zealots believe that the MMR vaccine causes autism, maternal rubella infection while the fetus is still in utero, as a cause of autism. Moreover, as P.Z. pointed out, no transient exposure or exposures to an external agent (such as a vaccine) is known to be able to produce such a consistent pattern of gene duplications and deletions in human cells like the sets detected in Pinto et al. Thus far, the preponderance of evidence points to primarily a genetic cause for autism and ASDs, and Pinto et al is another solid study supporting a genetic basis for autism. It also suggests potential cell signaling pathways that might be abnormal in autism and thus targets for therapeutic intervention. That’s all we can ask of such a study. It does not rule out an environmental component, although this study is not consistent with vaccines as an etiology of autism. More importantly, it has failed to provide any validating evidence for the frequently claimed abnormalities touted by autism biomeddlers, abnormalities allegedly targeted by all manner of quack nostroms, from chelation therapy to antioxidants to hyperbaric oxygen to “detoxification.”
Now remains the hard work of validating and replicating these findings and then figuring out which pathways can be targeted for therapy. What I’m afraid of now is that the quacks will look at the pathways identified by Pinto et al and try to come up with new pseudoscientific “biomed treatments” for autism based on these results, after (of course) “showing” how vaccine “injury” can create these same CNVs in children. It wouldn’t surprise me if I were to see this sort of “science” presented at Autism One next year.
On the other hand, maybe not. For quacks to try to use the results of Pinto et al to “treat” autism, they’d first have to accept the results of the study.
REFERENCE:
Pinto, D., Pagnamenta, A., Klei, L., Anney, R., Merico, D., Regan, R., Conroy, J., Magalhaes, T., Correia, C., Abrahams, B., Almeida, J., Bacchelli, E., Bader, G., Bailey, A., Baird, G., Battaglia, A., Berney, T., Bolshakova, N., Bölte, S., Bolton, P., Bourgeron, T., Brennan, S., Brian, J., Bryson, S., Carson, A., Casallo, G., Casey, J., Chung, B., Cochrane, L., Corsello, C., Crawford, E., Crossett, A., Cytrynbaum, C., Dawson, G., de Jonge, M., Delorme, R., Drmic, I., Duketis, E., Duque, F., Estes, A., Farrar, P., Fernandez, B., Folstein, S., Fombonne, E., Freitag, C., Gilbert, J., Gillberg, C., Glessner, J., Goldberg, J., Green, A., Green, J., Guter, S., Hakonarson, H., Heron, E., Hill, M., Holt, R., Howe, J., Hughes, G., Hus, V., Igliozzi, R., Kim, C., Klauck, S., Kolevzon, A., Korvatska, O., Kustanovich, V., Lajonchere, C., Lamb, J., Laskawiec, M., Leboyer, M., Le Couteur, A., Leventhal, B., Lionel, A., Liu, X., Lord, C., Lotspeich, L., Lund, S., Maestrini, E., Mahoney, W., Mantoulan, C., Marshall, C., McConachie, H., McDougle, C., McGrath, J., McMahon, W., Merikangas, A., Migita, O., Minshew, N., Mirza, G., Munson, J., Nelson, S., Noakes, C., Noor, A., Nygren, G., Oliveira, G., Papanikolaou, K., Parr, J., Parrini, B., Paton, T., Pickles, A., Pilorge, M., Piven, J., Ponting, C., Posey, D., Poustka, A., Poustka, F., Prasad, A., Ragoussis, J., Renshaw, K., Rickaby, J., Roberts, W., Roeder, K., Roge, B., Rutter, M., Bierut, L., Rice, J., Salt, J., Sansom, K., Sato, D., Segurado, R., Sequeira, A., Senman, L., Shah, N., Sheffield, V., Soorya, L., Sousa, I., Stein, O., Sykes, N., Stoppioni, V., Strawbridge, C., Tancredi, R., Tansey, K., Thiruvahindrapduram, B., Thompson, A., Thomson, S., Tryfon, A., Tsiantis, J., Van Engeland, H., Vincent, J., Volkmar, F., Wallace, S., Wang, K., Wang, Z., Wassink, T., Webber, C., Weksberg, R., Wing, K., Wittemeyer, K., Wood, S., Wu, J., Yaspan, B., Zurawiecki, D., Zwaigenbaum, L., Buxbaum, J., Cantor, R., Cook, E., Coon, H., Cuccaro, M., Devlin, B., Ennis, S., Gallagher, L., Geschwind, D., Gill, M., Haines, J., Hallmayer, J., Miller, J., Monaco, A., Nurnberger Jr, J., Paterson, A., Pericak-Vance, M., Schellenberg, G., Szatmari, P., Vicente, A., Vieland, V., Wijsman, E., Scherer, S., Sutcliffe, J., & Betancur, C. (2010). Functional impact of global rare copy number variation in autism spectrum disorders Nature DOI: 10.1038/nature09146